
Cancer-selective Mechanisms of Oncolytic Viruses
Abstract
Viruses that can be used for the treatment of cancer are referred to as oncolytic viruses. These viruses selectively infect cancer cells and induce cell death through a process of viral replication, proliferation, and budding. Diverse oncolytic viruses exist, and the genetic mechanisms of their cancer-selective killing strategies vary with the genetic diversity of each virus. Therefore, accurate information on viral genes and an understanding of cancer-selective killing mechanisms are required to study the treatment of cancer using oncolytic viruses. These require the storage and utilization of large amounts of data related to oncolytic viruses and cancer and studies based on enormous data sets using computer-based bioinformatics approaches. Such resources will aid in the development of novel cancer therapeutics using oncolytic viruses by providing the scientific basis for problem solving and decision making. In this study, we discuss the anti-cancer mechanisms of oncolytic viruses, classify them into four categories, and investigate the anti-cancer activities of different viruses. In addition, we evaluate the need for interdisciplinary research on oncolytic viruses by considering future research and therapy directions based on our understanding of molecular mechanisms, genetic mechanisms, and research achievements.
Keywords:
Oncolytic virus, cancer-selective mechanism, oncolysis, oncolytic virotherapy, immunotherapyIntroduction
Cancer is a major cause of death worldwide and should be resolved as soon as possible to improve human health and well-being. The conventional cancer treatment methods include surgical intervention, chemotherapy, and radiation therapy. In recent years, new treatments such as immunotherapy and gene therapy have also been used [1]. Studies on cancer treatment methods using viruses have also been performed [2]. Viruses that can be used for cancer treatment are referred to as oncolytic viruses, viruses that kill cancer cells. These viruses selectively infect tumor tissue and cause apoptosis through viral replication and proliferation within cancer cells. They show potential as novel cancer therapeutics by selectively killing cancer cells while causing no harm to normal tissues [3]. Oncolytic viruses can be divided into viruses that have selective infectivity towards cancer cells and those that can kill cancer cells through artificial manipulations and modifications geared toward the acquisition of selective infectivity. Viruses such as coxsackievirus, echovirus, reovirus, measles virus, poliovirus, and myxoma virus are included in the former group, and other viruses such as adenovirus, vesicular stomatitis virus, influenza virus, measles virus, vaccinia virus, and Newcastle disease virus belong to the latter group [4]. These cancer-killing viruses can be divided into DNA viruses and RNA viruses, and can function as ‘cancer killers’ through various anti-cancer mechanisms. Research on the development of cancer therapeutics with viruses is ongoing, and the anti-cancer effects of these virus-based therapeutic agents are being explored. Approximately 40 types of oncolytic viruses are being studied for their use as therapeutic agents [5]. Clinical trials of oncolytic viruses targeting various types of cancer are currently underway, and their anti-cancer effects are being demonstrated. Certain viruses are more commonly used due to their modification through genetic engineering.
Oncolytic viruses show diverse and complicated genetic mechanisms of killing cancer cells [6]. Table 1 shows features of major oncolytic viruses and their anticancer mechanisms. The diverse anti-cancer mechanisms of oncolytic viruses suggest that viruses can be applied as cancer therapeutic agents. However, our understanding of these mechanisms remains limited due to the lack of genetic evidence for the various mechanisms of cancer cell death by these viruses [7]. In recent years, studies have been performed to increase the efficiency of viral oncolysis and on genetic engineering methods to alter the cancer selectivity of viruses, and numerous studies have explored the design of novel oncolytic viruses. To construct oncolytic viruses that are safe and effective therapeutic agents, it is important to increase their specificity towards cancer cells, maximize their efficiency, and assess their safety [8]. Initially, an accurate understanding of the oncolysis strategies of each oncolytic virus is required. Studies of new mechanisms, the genes and proteins of the viruses and host cells involved in anticancer mechanisms, and characteristics of novel oncolytic virus candidates and genetic engineered viruses will be performed by studying the cancer-killing mechanisms used by each virus.
Cancer-selective infection: targeting cancer cell surface antigens
Although all oncolytic viruses have the ability to selectively kill cancer cells, different viruses employ different mechanisms. Figure 1 shows various strategies of oncolytic viruses that cause cancer-selective infectivity. In this article, we explore these mechanisms by classifying them into four categories and investigate the anti-cancer viral activities of each mechanism. We discuss the necessity of interdisciplinary research on oncolytic viruses as effective cancer therapeutics, considering future research directions.
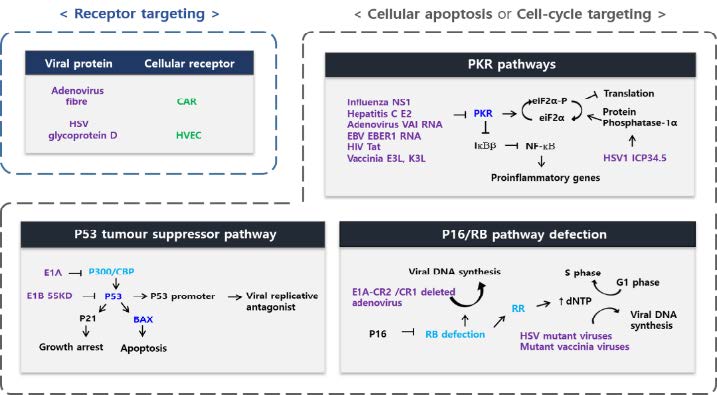
Various strategies for cancer-selective infectivity of oncolytic viruses.Oncolytic viruses that can selectively kill cancer cells have various strategies for targeting cancer cells due to different genetic and biomolecular characteristics. As a result of these strategies, various viral mechanisms for cancer-selective killing are used by each virus.
Viruses must first undergo attachment and penetration through cellular surface antigens to infect a host cell. Initially, the surface antigen protein of the virus attaches to specific receptor proteins on the surface of target host cells. The entrance of a virus into a host cell and transmission of the viral genome into its cytoplasm occurs through interactions between proteins of the virus and host cell. The virus enters the cell through viral membrane fusion in the case of enveloped viruses and by endocytosis in the case of non-enveloped viruses [9]. Viral attachment events during the infection process are the first step in the viral life cycle. This step is responsible for determining the host range of viruses due to the large variety of host cell surface proteins [10]. Therefore, cancer cells expressing a specific surface antigen may be targeted by specific viruses.
Several viruses, including mumps virus, reovirus, Newcastle disease virus, measles virus, and coxsackievirus, exhibit selective infectivity toward cancer cells, and other viruses, including adenovirus, vesicular stomatitis virus (VSV), herpes simplex virus (HSV), and vaccinia virus, can show selective infectivity towards cancer cells after genetic manipulation and transformation [8]. The first studied virus that showed potential as an oncolytic virus was an adenovirus [11]. Adenoviruses are enveloped DNA viruses in which the capsid protein surrounding the DNA genome contains ‘fibre’ protein. Because fibre protein can use coxsackie and adenovirus receptor (CAR) as a receptor protein, an adenovirus must be modified to create cancer-selective infectivity since CAR is expressed in both normal cells and cancer cells [12]. Measles virus has inherent cancer-selective infectivity, and can be genetically modified to show increased specificity towards cancer cells. Hemagglutinin (H) protein, an attachment protein present on the envelope of measles virus, can use CD46 as a receptor protein. Since CD46 is expressed on various cancer cells, the measles virus has a higher specific infectivity for cancer cells [13, 14]. In addition, many viruses show high infectivity toward cancer cells, including coxsackievirus, poliovirus, echovirus, and VSV [3, 15]. Thus, various viruses can exhibit specificity for cancer cells, obtain anticancer mechanisms through genetic manipulations, or exhibit a high infectivity rate toward cancer cells depending on the type of receptor protein used. Oncolytic viruses that are capable of targeting various cancer cells are being developed and targeted towards clinical trials.
Cancer-selective replication
Systematic infection by viruses requires viral genome replication using the replication machinery of host cells [16, 17]. Understanding the mechanisms that occur in cancer cells and the characteristics of the associated cancer micro-environment are needed to determine why these processes occur selectively in cancer cells and not in normal cells [18].
Several viruses show cancer-specific replication, including reovirus, which replicates exclusively in cancer cells [19]. Reovirus, an enveloped dsRNA virus, shows selective infectivity toward and a high replication rate in cancer cells, along with abnormal activity of the RAS pathway. Reolysin, which was developed using reovirus, has shown potential as a novel cancer therapeutic in clinical trials [20]. Vaccinia virus also has a high replication rate in cancer cells with abnormal epidermal growth factor receptor (EGFR) activity, which is relevant to the RAS pathway. VSV has a high replication rate in cancer cells due to their lower antiviral responses than normal cells. Defects or downregulation in the interferon pathway in cancer cells allow increased specificity of viruses towards cancer cells [21].
In addition to viruses with inherent cancer-selective replication, some viruses can obtain anticancer mechanisms through genetic engineering. Adenovirus can be used as an oncolytic virus after genetic modification; the modified version replicates and proliferates only in cancer cells with an abnormal p53-mediated cell cycle. The adenovirus E1B gene has been found to inactivate the tumor suppressor protein p53, so that adenovirus with a deleted E1 gene has a relatively high replication rate in cancer cells with deficient p53 because it no longer expresses E1B protein [22]. HSV with a deleted E1B gene selectively infects cancer cells; because the suppression of viral replication and proliferation do not work properly due to this deletion, apoptosis is induced [23]. Influenza virus infects pigs, birds, and humans, causing respiratory diseases. In addition, it is capable of being utilized as an oncolytic virus to perform a specific replication and multiplication in cancer cells. The non-structural protein 1 (NS1) protein produced by the influenza virus plays an important role in its pathogenicity by inhibiting PKR-mediated antiviral responses. Therefore, cancer-specific apoptosis can be induced after viral replication in normal cells with normal PKR activity, and selective viral replication can occur in cancer cells by deleting the NS1 protein from viruses [24].
In summary, genetic engineering techniques have been used to create numerous viruses that show cancer-selective replication and proliferation as well as improved anti-cancer effectiveness. New mechanisms of cancer-specific replication have been identified, likely with more to come. Studies to explore various viruses as cancer therapeutics based on these mechanisms will serve as foundations for novel cancer therapeutics that utilize oncolytic viruses.
Targeting cancer microenvironment
Differences exist between themicroenvironments of various cancer cells, and viruses targeting the cancer microenvironment can be used for cancer-specific killing.
Reovirus, a non-enveloped virus that is sensitive to cancer microenvironments, typically infects cells of digestive organs. Proteases are present at high levels in digestive organs and can convert non-infectious reovirus into infectious viral particles. Proteases are commonly overexpressed in the cancer microenvironment, and reovirus can act as an infectious particle by activating a protease. For this reason, reovirus has a higher infection rate of cancer cells than of normal cells, and eventually causes cancer-selective apoptosis [25].
Use of the cancer microenvironment to increase selectivity towards cancer cells can also be applied to measles virus and Newcastle disease virus. The fusion (F) protein of measles virus mediates the fusion of viruses with the cell membrane; it is a surface protein that functions during virus attachment and penetration into the host cell. F protein typically exists as an inactive precursor, and the fusion peptide is exposed when cleaved by furin. Therefore, it is possible to construct a virus that is converted to its active form only in the protease-rich cancer microenvironment by altering the F protein [26]. Proteases such as matrix metalloproteinase 2 (MMP2) are expressed at high levels in various types of cancer cells. Therefore, the infection rate of measles virus towards cancer cells can be increased by activating viral pathogenicity in the vicinity of cancer cells through the modification of the F protein to contain an MMP2 cleavage site in place of the furin site. Because Newcastle disease virus also contains the F protein, it is possible to increase the viral infection rate by targeting the cancer microenvironment in the same way as the measles virus [26]. To target viruses to the cancer microenvironment using modification and replacement of the cleavage site of the viral fusion protein, studies on how to increase the specificity of viruses toward cancer cells or the cancer microenvironment are required.
Immunotherapy using oncolytic viruses
Conventional anticancer therapies typically include surgical intervention, radiation therapy, or chemotherapy, depending on the condition of individual patients. In addition to these traditional therapies, studies on immunotherapy using the patient's immune system and the introduction of genes involved in cancer-related mechanisms are currently underway, and the results of several recent clinical trials have been successful [1, 2].
Cancer therapy using oncolytic viruses is a novel cancer treatment method. The cancer-selective killing mechanisms of certain viruses, as well as the results of substitution, introduction, or deletion of viral genes through genetic modification, suggest that viral gene therapies can be used for cancer treatment. Gene therapy using oncolytic viruses does not have as high of cure rate as conventional anticancer therapies. However, further studies are warranted because this type of gene therapy causes fewer side effects than conventional chemotherapy. Anticancer immune therapy shows efficacy with remarkably low side effects because the individual patient’s own cells are used for the treatment, unlike conventional anti-cancer agents. Immune cells such as natural killer (NK) cells, dendritic cells, B cells, and T cells are used in cancer immunotherapy and can be combined with existing anticancer therapeutic agents [27]. Oncolytic viruses can also be used for anticancer immunotherapy. It is possible to induce apoptosis in cancer cells indirectly by stimulating anticancer immune responses through oncolytic viruses [28]. Oncolytic viruses used in cancer immunotherapy function as vectors for the expression of genes that stimulate viral-mediated immune responses, and viruses such as herpes simplex virus-1 (HSV-1), vaccinia virus, and adenovirus have these anticancer mechanisms [29]. HSV-1 can be used in anticancer immunotherapy after genetic manipulation. OncoVEX GM-CSF, an attenuated HSV-1 created by genetic modification, has been evaluated in clinical trials in patients with metastatic melanoma [30]. The vaccinia virus line JX-594, which has been modified to express GM-CSF, was shown to have anticancer effects in patients with liver cancer and melanoma [31]. GM-CSF can also be introduced into adenovirus, resulting in anticancer effects. Such lines have been shown to stimulate the immune system against prostate cancer through experiments in hamster. It is difficult to predict the responses and effects of anticancer immune therapy with oncolytic viruses, and extensive examination of appropriate models is needed. Therefore, studies on the molecular biology of the virus and immune system of the host cell are required.
Conclusion
The development and use of various cancer treatments require the genetic analysis of oncolytic viruses with potential as advanced cancer therapeutics and studies on their anticancer mechanisms. Mouse models, cancer cell lines, and clinical experiments in cancer patients have been used in most studies on oncolytic viruses. However, these studies are time-consuming and costly, and can have safety risks due to the biological characteristics of the viruses. Thus, novel methods to address these problems are required. Bioinformatics studies will provide a novel perspective in oncolytic virus studies. Above all, understanding the complex mechanisms of the various viruses is required to study their complex cancer-selective mechanisms. This will require the construction of databases and analytical tools that enable researchers to utilize viral sequence information and other biological data. Figure 2 shows the utilization and expected achievements of database-based bioinformatics approaches in studies of oncolytic viruses.
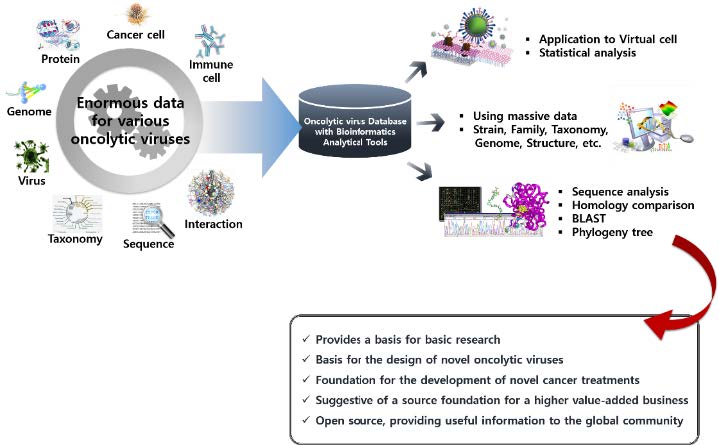
Use of bioinformatics for studies of oncolytic viruses.An analysis of cancer-selective killing mechanisms of viruses having potential as novel cancer therapeutics by constructing a specialized database integrating enormous amounts of data for oncolytic viruses is shown. Based on the constructed database, the development of an integrated bioinformatics technique facilitating the analysis and prediction of biological mechanisms can be undertaken.
Correlation analysis of genetic properties and the anticancer mechanisms of each virus will be studied using integrated databases, and candidate viruses for clinical trials will be evaluated using systematic analysis of oncolytic viruses based on studies of genetic information and biomolecular mechanisms. Studies of the anti-cancer mechanisms of oncolytic viruses will benefit from the effects of enhanced competitiveness of national medical technology by contributing to the development of novel cancer therapeutics through the analysis of genetic variables in viruses and host cells. From an international perspective, studies on cancer have great value to society in that they have high value if state-of-the-art therapeutics are made available, with global demand. More diverse data will be accumulated through future studies. It is believed that the convergence of interdisciplinary research and the application of findings to relevant studies will be accomplished by sharing information and promoting communication among researchers during the construction of an integrated database. Collaborative studies on oncolytic viruses performed in various academic fields, including bioinformatics, are expected to provide novel insights into the development of strategies for cancer treatment.
In this review, various cancer-selective oncolysis mechanisms of oncolytic viruses that have potential as cancer therapeutics were explored by classification according to individual viral genetic and molecular features, and the anti-cancer activities of each virus and research trends were investigated. In addition, the necessity of interdisciplinary research including bioinformatics was evaluated for studies on oncolytic viruses. Cancer is the leading cause of death in Korea, and studies on the development of novel cancer therapeutics are important for public health. Because social and economic losses have gradually increased due to a steady increase in the incidence of cancer and mortality, novel cancer therapeutics are required. It is believed that bioinformatics approaches capable of scientific analysis based on the integration and processing of a large amount of data will provide novel solutions to replace conventional research methods. Newly derived research results based on interdisciplinary studies including bioinformatics approaches will allow for the creation of novel information and applications. As a result, the groundwork for future studies on cancer treatment is available and is expected to provide a scientific basis for the practical application of advanced cancer treatment methods.
Acknowledgments
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korean government (MEST 2012008344). This research was also supported by the Bio & Medical Technology Development Program of the National Research Foundation (NRF) funded by the Korean government (MEST 2012M3A9D1054622). Support from Korea Institute of Science and Technology Information (KISTI K-13-L01-C02-S04) is gratefully acknowledged. This work was partly supported by CMB.
References
-
Cross, D., Burmester, JK., Gene therapy for cancer treatment: past, present and future, Clinical Medicine & Research, (2006), 4(3), p218-227.
[https://doi.org/10.3121/cmr.4.3.218]

-
Alemany, R., Viruses in cancer treatment, Clinical and Translational Oncology, (2013), 15(3), p182-188.
[https://doi.org/10.1007/s12094-012-0951-7]

-
Kelly, E., Russell, SJ., History of oncolytic viruses: genesis to genetic engineering, Molecular Therapy, (2007), 15(4), p651-659.
[https://doi.org/10.1038/sj.mt.6300108]

-
Nemunaitis, J., Oncolytic viruses, Investigational new drugs, (1999), 17(4), p375-386.
[https://doi.org/10.1023/A:1006334404767]

-
Russell, SJ., Peng, KW., Bell, JC., Oncolytic virotherapy, Nature biotechnology, (2012), 30(7), p1-13.
[https://doi.org/10.1038/nbt.2287]

-
Hawkins, LK., Lemoine, NR., Kirn, D., Oncolytic biotherapy: a novel therapeutic platform, The lancet oncology, (2002), 3(1), p17-26.
[https://doi.org/10.1016/S1470-2045(01)00618-0]

-
Davis, JJ., Fang, B., Oncolytic virotherapy for cancer treatment: challenges and solutions, The journal of gene medicine, (2005), 7(11), p1380-1389.
[https://doi.org/10.1002/jgm.800]

-
Chiocca, EA., Oncolytic viruses, Nature Reviews Cancer, (2002), 2(12), p938-951.
[https://doi.org/10.1038/nrc948]

-
Chazal, N., Gerlier, D., Virus Entry, Assembly, Budding, and Membrane Rafts, Microbiology and molecular biology reviews, (2003), 67(2), p226-237.
[https://doi.org/10.1128/MMBR.67.2.226-237.2003]

-
Dimitrov, DS., Virus entry: molecular mechanisms and biomedical applications, Nature Reviews Microbiology, (2004), 2(2), p109-122.
[https://doi.org/10.1038/nrmicro817]

-
Garber, K., China approves world’s first oncolytic virus therapy for cancer treatment, Journal of the National Cancer Institute, (2006), 98(5), p298-300.
[https://doi.org/10.1093/jnci/djj111]

-
Bewley, MC., Springer, K., Zhang, YB., Freimuth, P., Flanagan, JM., Structural analysis of the mechanism of adenovirus binding to its human cellular receptor, CAR, Science, (1999), 286(5444), p1579-1583.
[https://doi.org/10.1126/science.286.5444.1579]

-
Fielding, AK., Measles as a potential oncolytic virus, Reviews in medical virology, (2005), 15(2), p135-142.
[https://doi.org/10.1002/rmv.455]

-
Anderson, BD., Nakamura, T., Russell, SJ., Peng, KW., High CD46 receptor density determines preferential killing of tumor cells by oncolytic measles virus, Cancer research, (2004), 64(14), p4919-4926.
[https://doi.org/10.1158/0008-5472.CAN-04-0884]

-
Bergman, I., Whitaker-Dowling, P., Gao, Y., Griffin, JA., Watkinse, SC., Vesicular stomatitis virus expressing a chimeric Sindbis glycoprotein containing an Fc antibody binding domain targets to Her2/neu overexpressing breast cancer cells, Virology, (2003), 316(2), p337-347.
[https://doi.org/10.1016/j.virol.2003.07.010]

-
Sieczkarski, SB., Whittaker, GR., Viral Entry, In Membrane Trafficking in Viral Replication, Springer Berlin Heidelberg, (2005), p1-23.
[https://doi.org/10.1007/3-540-26764-6_1]

-
Miller, S., Krijnse-Locker, J., Modification of intracellular membrane structures for virus replication, Nature Reviews Microbiology, (2008), 6(5), p363-374.
[https://doi.org/10.1038/nrmicro1890]

-
Mohr, I., To replicate or not to replicate: achieving selective oncolytic virus replication in cancer cells through translational control, Oncogene, (2005), 24(52), p7697-7709.
[https://doi.org/10.1038/sj.onc.1209053]

-
Norman, KL., Lee, PW., Not all viruses are bad guys: the case for reovirus in cancer therapy, Drug discovery today, (2005), 10(12), p847-855.
[https://doi.org/10.1016/S1359-6446(05)03483-5]

-
Norman, KL., Hirasawa, K., Yang, AD., Shields, MA., Lee, PW., Reovirus oncolysis: The Ras/RalGEF/p38 pathway dictates host cell permissiveness to reovirus infection, Proceedings of the National Academy of Sciences of the United States of America, (2004), 101(30), p11099-11104.
[https://doi.org/10.1073/pnas.0404310101]

-
Stojdl, DF., Lichty, B., Knowles, S., Marius, R., Atkins, H., Sonenberg, N., Bell, JC., Exploiting tumorspecific defects in the interferon pathway with a previously unknown oncolytic virus, Nature medicine, (2000), 6(7), p821-825.
[https://doi.org/10.1038/77558]

- Rothmann, T., Hengstermann, A., Whitaker, NJ., Scheffner, M., Hausen, HZ., Replication of Onyx-015, a potential anticancer adenovirus, is independent of p53 status in tumor cells, Journal of virology, (1998), 72(12), p9470-9478.
-
Heise, C., Sampson-Johannes, A., Williams, A., Mccormick, F., Von Hoff, DD., Kirn, DH., ONYX-015, an E1B gene-attenuated adenovirus, causes tumor-specific cytolysis and antitumoral efficacy that can be augmented by standard chemotherapeutic agents, Nature medicine, (1997), 3(6), p639-645.
[https://doi.org/10.1038/nm0697-639]

-
Garcı́, A., Egorov, A., Matassov, D., Brandt, S., Levy, DE., Durbin, JE., Palese, P., Muster, T., Influenza A Virus Lacking the NS1 Gene Replicates in Interferon-Deficient Systems, Virology, (1998), 252(2), p324-330.
[https://doi.org/10.1006/viro.1998.9508]

-
Klenk, HD., Garten, W., Host cell proteases controlling virus pathogenicity, Trends in microbiology, (1994), 2(2), p39-43.
[https://doi.org/10.1016/0966-842X(94)90123-6]

-
Nagai, Y., Klenk, HD., Rott, R., Proteolytic Cleavage of the Viral Glycoproteins and Its Significance for the Virulence of Newcastle Disease Virus, Virology, (1976), 72(2), p494-508.
[https://doi.org/10.1016/0042-6822(76)90178-1]

-
Bhat, R., Dempe, S., Dinsart, C., Rommelaere, J., Enhancement of NK cell antitumor responses using an oncolytic parvovirus, International Journal of Cancer, (2011), 128(4), p908-919.
[https://doi.org/10.1002/ijc.25415]

-
Kirn, DH., Thorne, SH., Targeted and armed oncolytic poxviruses: a novel multi-mechanistic therapeutic class for cancer, Nature Reviews Cancer, (2009), 9(1), p64-71.
[https://doi.org/10.1038/nrc2545]

-
Hernández-Alcoceba, R., Recent advances in oncolytic virus design, Clinical and Translational Oncology, (2011), 13(4), p229-239.
[https://doi.org/10.1007/s12094-011-0647-4]

-
Kaufman, HD., Kim, DW., DeRaffele, G., Mitcham, J., Coffin, RS., Kim-Schulze, S., Local and distant immunity induced by intralesional vaccination with an oncolytic herpes virus encoding GMCSF in patients with stage IIIc and IV melanoma, Annals of surgical oncology, (2010), 17(3), p718-730.
[https://doi.org/10.1245/s10434-009-0809-6]

-
Lee, JH., Roh, MS., Lee, YK., Kim, MK., Han, JY., Park, BH., Trown, P., Kirn, DH., Hwang, TH., Oncolytic and immunostimulatory efficacy of a targeted oncolytic poxvirus expressing human GM-CSF following intravenous administration in a rabbit tumor model, Cancer gene therapy, (2010), 17(2), p73-79.
[https://doi.org/10.1038/cgt.2009.50]

